\\// Home \\//Genetics Theory \\// Chicken Mutations\\// Chromosome Linkages\\//
\\//Chicken Variety Genotypes\\//
Genetics Calculators\\//
\\// References/Links\\// About / Contact Us\\//
Old URL: http://home.ezweb.com.au/~kazballea/genetics/theory.html
Last updated: 29 May, 2007
* Most of the photos on this website originated from "Feathersite",
and "Old English Bantam Club of America" websites, and have been given with
permission by the website administrators.
© Copyright of images belongs to cited photography contributors.
-----------------------------------------------------------------------------
* ![]() Click here for "Print Version"
Click here for "Print Version"
* ![]() Download (..rignt click_Save Target As..) "PDF Version (724 KB) "
Download (..rignt click_Save Target As..) "PDF Version (724 KB) "
-----------------------------------------------------------------------------
| Genotype: | The genetic makeup of the organism |
| Phenotype: | The physical appearance of the organism |
| Sexual Dimorphism: | “Sexual dimorphism” includes differing colour/pattern forms between the two sexes of a species, i.e. not restricted to physical body traits. The gender colour/pattern differences in the wild type – Red Jungle Fowl (black breasted red roosters, salmon breasted hens, etc) are two alternate colour/pattern forms, the particular form expressed dependent on gender. |
| P1 | Parent 1 (in first cross) |
| F1 F2...... | First Filial, Second Filial (1st Generation, etc) |
| BC1.... etc: | Back Cross 1 (cross F1 back to Parent, etc) |
| Gene | A unit of hereditary , a section of DNA found on a chromosome that codes for a particular protein |
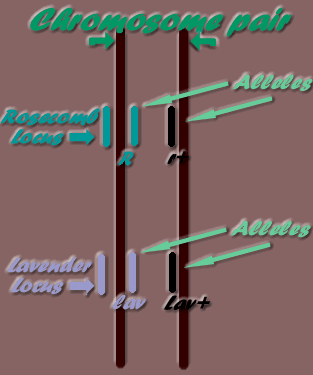
| Locus (Loci - plural) | The location of an Allele on the Chromosome |
| Allele | One of two alternate forms of a gene that has the same locus on homologous chromosomes |
| Chromosome | A threadlike body in the cell nucleus that carries the genes in a linear order |
| Homozygous | Where alleles of a locus on homologous chromosomes are the same |
| Heterozygous | Where alleles of a locus on homologous chromosomes are different |
| Hemizygous | A genetic locus present in one copy only. Of females, in reference to the only one allele at each locus on the single Z chromosome (only one allele possible, as only one Z chromosome in females, therefore not heterozygous or homozygous as no chromosome pair). |
| Linkage Map Unit, (centiMorgan) | 1 map unit, or 1 centiMorgan (cM) is equal to 1% recombination |
Inheritance Mode Types: | |
| Dominant: | Of genes; producing the same phenotype whether its allele is identical or dissimilar |
| Recessive: | Of genes; producing its characteristic phenotype only when its allele is identical |
| Incompletely dominant/recessive: | When heterozygous, giving an intermediate phenotype |
| Co-dominant: | When heterozygous, expressing both alleles |
| Sex-linked dominant/recessive: | Of genes; on the Z chromosome |
| Linkages: | Where loci do not segregate independently |
| Sex-Limited: | Where both genders carrying the gene, but gene expression is with one gender only (eg, egg-shell colour can only be expressed in hens) |
| Sex-Influenced: | Where a gene may appear dominant with one gender, recessive in another, ie gene expression is different between genders when alleles heterozygous |
| Autosomal: | Any chromosome that is not a sex chromosome; appear in pairs in body cells |
Others: | |
| Pleiotrophy: | Multiple traits expressed by a single gene |
| Polygenic trait: | Multiple genes giving accumulative effect on trait expression |
| Epistasis | Gene expression affected by a gene from another locus |
| Hypostasis: | The converse of epistasis, applied to the gene pair hidden by the epistatic gene pair |
| Penetrance: | The proportion of individuals of a specified genotype that express the expected phenotype |
| Expressivity: | The range of phenotypes expressed by a given genotype |
| Homologous Chromosomes: | Corresponding or similar in position |
| Multiple Alleles : | Where more than one mutation has occurred on a specific locus |
| Diploid: | An organism or cell having two sets of chromosomes |
| Pheomelanin: | Pigments that account for wild type red colour |
| Eumelanin: | Pigments that account for wild type black colour |
Genetics Theory:
Chickens have 78 chromosomes. They are diploid animals, therefore the body cell chromosomes are grouped together in pairs- 39. For example, a chicken will have two Chromosome 1's, two chromosome 2's, two chromosome 3's, etc. The exception is the sex-chromosomes, Z and W, where roosters have two Z chromosomes and hens have only one Z chromosome, plus one W chromosome.
The following diagram may be useful in understanding the genetics terms:
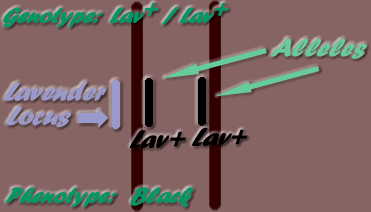
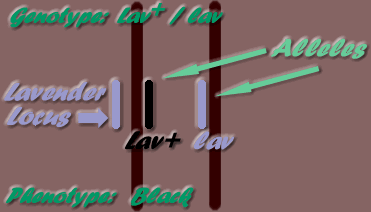
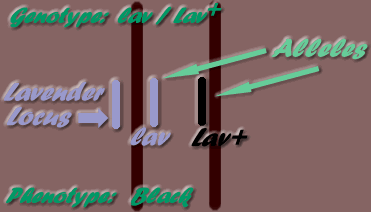
The position along a chromosome where a gene may be found, is its locus. "Alleles" are the alternate genes possible on a specific locus. For example, the lavender gene (lav) may only be found on the Lavender locus. As only one mutation has been known to occur at this locus, there is a total of two alleles possible, ie the lav mutation and the wild type allele - Lav+ (wild type alleles are identified by the plus symbol: + ). Therefore, the possible locus allele combinations on the two homologous chromosomes are:
- - Lav+ / Lav+
- - Lav+ / lav
- - lav / Lav+
- - lav / lav

Sometimes multiple alleles have mutated on a given locus. That is, there may be a list of alleles available, not just two (ie, not just one mutation & the wild type allele). For example, E, ER, ER- Fayoumi, eWh, e+, eb, es, ebc, ey, eq alleles are all on the one locus (E locus). But only two alleles are physically possible in a diploid individual, at any given locus, ie, an individual may have ER/e+, or E/ER, or eb /E, etc, but only two alleles from the E Series.

----------------------------------------------------------------------------
As mentioned previously, sex chromosomes are different to the above autosome pairs, and different again between roosters (2 Z chromosomes) and hens (W & Z chromosomes). The following diagrams may be useful in understanding the differences between male & female sex chromosome pairs:
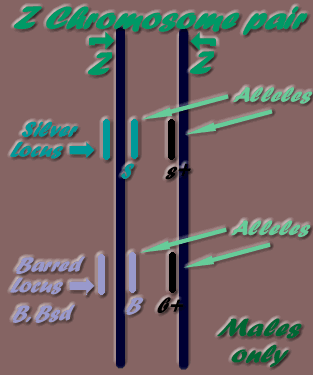
* Z chromosome pair- Males only
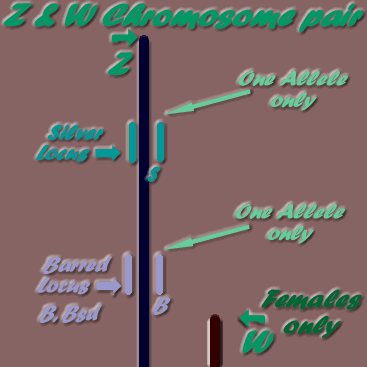
* Z & W chromosome pair- Females only
Males may have two alleles the same- homozygous, or if two different alleles- heterozygous (two different alleles on the one locus), but as females can only have one allele, as only one Z chromosome present, they are referred to as hemizygous.
![]()
-----------------------------------------------------------------------------
Wild type (Red Jungle Fowl - Gallus gallus)
The "Wild Type" is what the wild species population appears like. For the domestic fowl, Jaap and Hollander (1954) suggested that the wild type Red Jungle Fowl (Gallus gallus) be taken as the standard from which all deviations (or mutations) are measured (Carefoot, 1985).
References:
Carefoot, W.C. (1985) Creative Poultry Breeding.
Jaap, R.G. and Hollander, W.F. (1954) Wild type as standard in poultry genetics. Poultry Sci. 34:389-395.

*photo of Red Jungle Fowl
(Photograph from Feathersite.com)
Examples of Domestic Fowl breeds/varieties similar in colour/pattern are Brown Leghorns, Partridge (Light Brown) Dutch Bantams, Black Breasted Red- Partridge Old English Games, etc. The following is a photo of Black Breasted Red- Partridge Rosecombs.

* Black Breasted Red- Partridge Rosecombs
(Breeder, & Photography by Katherine Plumer -www.rosecomb.com website)
![]()
-----------------------------------------------------------------------------
Pheomelanin and Eumelanin Pigment Areas
"Pheomelanin" equals the red pigment areas, "Eumelanin" equals the black pigment areas. Some genes affect the pheomelanin areas only, eg., if a bird has the cream- ig gene, the red is diluted to cream, if silver- S mutation present, the pheomelanin red areas are changed to silver -white, if mahogany- Mh, the red is darkened to deep mahogany, etc. Some genes affect the eumelanin areas only, eg., if a bird has the blue, dominant white, dun, smoky, etc genes, the eumelanin is diluted to these colours. Other genes affect both eumelanin and pheomelanin, eg the Lavender- lav gene dilutes the eumelanin- black to a pale lavender shade, and the pheomelanin- red to a pale straw-cream shade. The Recessive White gene changes all pigmented plumage (pheomelanin and eumelanin) to white, regardless of other mutations in the genotype.

*Pheomelanin (red) & eumelanin (black) in wild type colour/pattern (e+)
(Breeder, & Photography by Katherine Plumer -www.rosecomb.com website)
Examples of Pheomelanin and Eumelanin mutations
*put mouse curser over pigment colours (to the left), to see how the mutations change the wild-type
|
* (Photographs from the OEGBCA website, & CJR - Cream Light Brown Dutch Bantams)
![]()
-----------------------------------------------------------------------------
Inheritance Modes, using Punnett Squares:
The three main inheritance modes are:
|
* Images used in the following punnet squares are from Feathersite, and the OEGBCA website.
Dominant / Recessive Inheritance
This inheritance mode is where one dose only of a dominant gene is needed for expression to occur (ie heterozygotes & homozygotes), but two doses are needed for expression of a recessive gene (ie homozygotes only express the gene). The example given below is of Lavender (lav- recessive gene) bird paired with a Black (Lav+ - dominant - wild type allele) bird.
* First Generation cross results, using Lavender & Black parents:
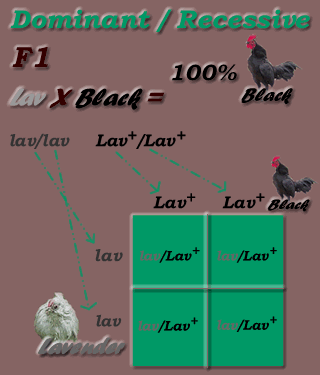
*Dominant / Recessive Inheritance Mode, F1 generation
Genotypic Percentages = 100% Lav+/lav
Phenotypic Percentages = 100% black
----------------------------------------------
* Second Generation cross (inbreed offspring - F1 X F1) results:
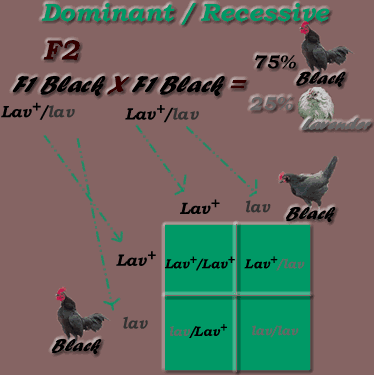
*Monohybrid Cross: Dominant / Recessive Inheritance Mode, F2 generation
Genotypic Ratio = 1:2:1 (1 Lav+/ Lav+ : 2 Lav+/lav : 1 lav/lav)
Phenotypic Ratio = 3:1 (3 black, 1 lavender)
------------------------------------
Breeding homozygous Black (Lav+/ Lav+) to homozygous Black (Lav+/ Lav+) will produce all homozygous Black (Lav+/ Lav+). Breeding homozygous Lavender (lav/lav) to homozygous Lavender (lav/lav) will produce all homozygous Lavender (lav/lav).
Other dominant/ recessive breeding combination examples are:
- Black, carrying Lavender Gene (Lav+/lav) paired to homozygous Black (Lav+/Lav+).
This produces 100% Black phenotype, but 50% are carrying the Lavender gene:
- 50% Lav+/ Lav+ (homozygous Black)
- 50% Lav+/lav (Black, carrying lavender)
- Black, carrying Lavender Gene (Lav+/lav) paired to Lavender (lav/lav).
This produces 50% Black phenotype, 50% Lavender phenotype. All the Blacks are carrying the Lavender gene:
- 50% Lav+/lav (Black, carrying lavender)
- 50% lav/lav
(Lavender)
---------------------------------------------------------------
Incomplete Dominant Inheritance
In the dominant/ recessive example, the Black, carrying lavender (heterozygote) bird is not distinguishable from the homozygous Black (not carrying lavender gene), as the black (wild type- Lav+) gene is completely dominant to the lavender gene. With Incompletely Dominant inheritance, the heterozygote gives an intermediate phenotype. For example, a splash (whitish- pale blue) bird (two doses of the Blue gene- Bl/Bl) paired to a black bird (no blue genes - bl+/bl+ -wild type) will produce an intermediate shade between the whitish/splash and black, ie- blue. The following is a punnet square example of this splash to black cross:
* First Generation cross results, using Splash & Black parents:
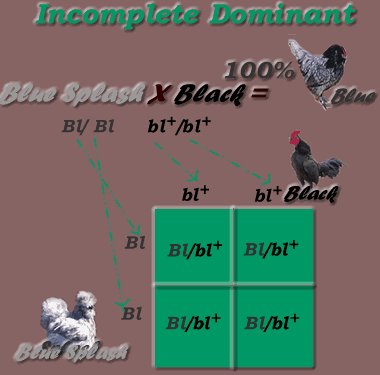
* Incomplete Dominant Inheritance Mode, F1 generation
Genotypic Percentages = 100% Bl/bl+
Phenotypic Percentages = 100% Blue
-------------------------------------
Breeding these Blue (F1) birds together will produce the following offspring percentages:
- Blue (Bl/bl+) paired to Blue (Bl/bl+):
This produces 25% Black phenotype, 50% Blue phenotype, 25% Splash phenotype:
- 25% bl+/bl+ (Black)
- 50% Bl/bl+ (Blue)
- 25% Bl/Bl (Splash)
* Second Generation cross (inbreed offspring -Blue F1 X Blue F1) results:
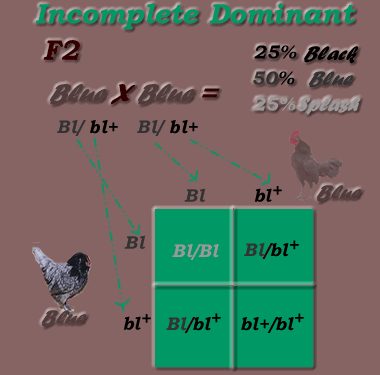
*Monohybrid Cross: Incomplete Dominant / Incomplete Recessive Inheritance Mode, F2 generation
Genotypic Ratio = 1:2:1 (1 Bl/B1 : 2 Bl/bl+ : 1 bl +/ bl +)
Phenotypic Ratio = 1:2:1 (1 splash, 2 blue, 1 black)
-------------------------------------
Breeding homozygous Black (bl+/bl+) to homozygous Black (bl+/bl+) will produce all homozygous Black (bl+/bl+). Breeding homozygous Blue-Splash (Bl/Bl) to homozygous Blue-Splash (Bl/Bl) will produce all homozygous Blue-Splash (Bl/Bl).
Other incompletely dominant/ incompletely recessive breeding combination examples are:
- Black (bl+/bl+) paired to Blue (Bl/bl+).
This produces 50% Black phenotype, 50% Blue phenotype:
- 50% bl+/bl+ (Black)
- 50% Bl/bl+ (Blue)
- Splash (Bl/Bl) paired to Blue (Bl/bl+).
This produces 50% Splash phenotype, 50% Blue phenotype:
- 50% Bl/Bl (Splash)
- 50% Bl/bl+ (Blue)
-----------------------------------------------------------
Sex-Linked Inheritance
Sex-Linked genes are genes on the Z chromosome. Roosters have two Z chromosomes and hens have only one Z chromosome, plus one W chromosome. As the hens only have one sex-linked allele for any given locus, both dominant and recessive sex-linked genes are expressed with just one gene (as hemizygous).
If the sex-linked recessive allele is homozygous in the male, and the sex-linked dominant allele is found in the female, all offspring males will inherit the sex-linked dominant gene from the mother, and all offspring females will inherit the sex-linked recessive gene from the fathers. The following is a punnet square example of a sex-linked cross, where all offspring females are the colour of the father and all offspring males are the colour of the mother. The recipricol cross (ie parent males with sex-linked dominant genes, parent females with sex-linked recessive gene) doesn't work the same, as all offspring will have the dominant gene.
* First Generation cross results, using Gold male & Silver female parents:
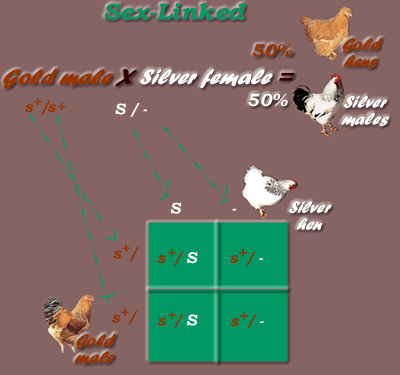
*Sex-Linked Dominant / Recessive Inheritance Mode, F1 generation
Genotypic Ratio = 1:1 (1 s+/ S : 1 s+ /-)
Phenotypic Ratio = 1:1 (1 silver- all males, 1 gold. all females)
-----------------------------------------
* Second Generation cross (inbreed offspring -Silver F1 X Silver F1) results:
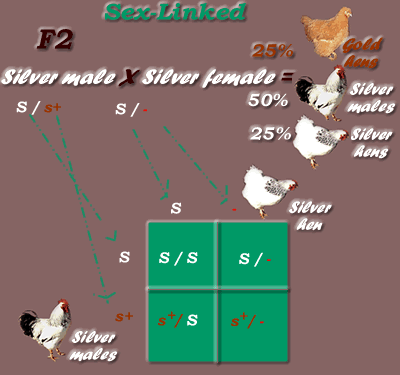
*Sex-Linked Dominant / Recessive Inheritance Mode, F2 Generation
Genotypic Ratio = 1:1:1:1 (1 S/S : 1 S/- : 1 s+/ S : 1 s+ /-)
Phenotypic Ratio = 1:2:1 (1 silver females, 2 silver males, 1 gold females) or
Colour only Phenotypic Ratio = 1:3 (1 gold, 3 silver)
-------------------------------------
Breeding homozygous gold (s+/s+) to hemizygous gold (s+/-) will produce all homozygous/hemizygous gold (s+/s+ or s+/-). Breeding homozygous Silver (S/S) to hemizygous Silver (S/-) will produce all homozygous/hemizygous Silver (S/S or S/-).
Other sex-linked breeding combination examples are:
- Silver, carrying Gold (S/s+) rooster paired to Gold (s+/-) hen.
This produces 50% Silver phenotype, 50% Gold phenotype, in both genders:
- 25% S/s+ (Silver males, carrying gold)
- 25% S/- (Silver females)
- 25% s+ /s+ (Gold males)
- 25% s+/- (Gold females)
- Silver (S/S) rooster paired to Gold (s+/-) hen.
This produces 100% Silver phenotype in both genders:
- 50% S/s+ (Silver males, carrying gold)
- 50% S/- (Silver females)
------------------------------------------------------------------
Dihybrid and other Multiple Mutation Crosses
When more than one mutation is in the equation, punnett squares become cumbersome. This is where the "Product Rule" comes in to play.
Product Rule:
prob(a and b) = p(a)p(b)
*which, in simple terms, is determined by finding the probability of each segregation that may occur in a pairing, then multiplying the probabilities together. For example, if it is determined that 1/4 of offspring will be Lavender, and 1/4 of offspring will be Mottled, it's a simple matter of multiplying the probabilities together to find the probability of segregating both Lavender and Mottled in an individual:
1/4 (Lavender) X 1/4 (Mottled) = 1/16 Lavender Mottled
Two mutations |
|
First Cross: P1 (Lavender Mottled) X P2 (Black) |
|
lav/lav mo/mo |
Lav+/Lav+ Mo+/Mo+ |
*Two Mutations, Dominant / Recessive Inheritance Mode, F1 generation
Genotypic Percentages = 100% Lav+/lav Mo+/mo
Phenotypic Percentages = 100% black
Dihybrid Cross: Two mutations |
|
|---|---|
Second Cross: F1 (black dihybrid) X F1 (black dihybrid) |
|
Lav+/lav Mo+/mo |
Lav+/lav Mo+/mo |
Gametes: Lav+ Mo+ , Lav+ mo, lav Mo+, lav mo Dihybrid Phenotypic Inheritance Ratio: 9:3:3:1 |
|
3/4 X 3/4 = 9/16 |
|
3/4 X 1/4 = 3/16 |
|
1/4 X 3/4 = 3/16 |
|
1/4 X 1/4 = 1/16 |
|
*Dihybrid Cross: Dominant / Recessive Inheritance Mode, F2 generation
Phenotypic Ratio = 9:3:3:1 (9/16 black, 3/16 black mottled, 3/16 lavender, 1/16 lavender mottled)
Dihybrid Genotypes:
 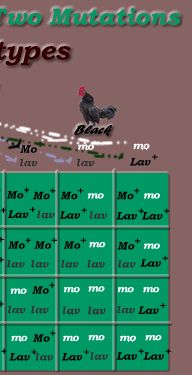 |
|---|
*Dihybrid Cross: Dominant / Recessive Inheritance Mode, F2 generation
Gametic Ratio = 1.1.1.1 (ie 1/4 Mo+ Lav+, 1/4 Mo+, lav, 1/4 mo lav, 1/4 mo Lav+)
Genotypic Ratio = 1:2:1: 2:4:2: 1:2:1
- 1 Mo+/Mo+ Lav+/ Lav+
- 2 Mo+/Mo+ Lav+/ lav
- 1 Mo+/Mo+ lav/lav
- 2 Mo+/mo Lav+/ Lav+
- 4
Mo+/mo Lav+/ lav
-2
Mo+/mo lav / lav
- 1 mo/mo Lav+/ Lav+
- 2 mo/mo
Lav+/ lav
- 1 mo/mo lav/lav
----------------------------------------------------------
Linkages
- Linkages occur where loci do not assort independently. Generally loci do assort independently, with crossover events common (eg, the recessive white gene assorts independently from the Pea Comb gene, as no linkage between their loci). For there to be a linkage between loci, they must be on the same homologous chromosome and in close proximity (less than 50 map units). Linked loci crossover less than 50 percent, as once they reach this distance, they assort independently. It is not uncommon for loci to assort independently of others even if on the same chromosome, as chromosomes can be very long and obtain many loci. For example, the S- Silver locus is not linked with the B - Barring locus, as over 50 map units apart on the Z chromosome.
The distance between the loci is measured in map units (centiMorgans) and is determined by the following equation:
Number of Recombinants |
||
100 X |
-------------------------------- |
= % of recombinants |
Number of Offspring |
||
* 1 map unit, or 1 centiMorgan is equal to 1% recombination
* The smaller the percentage of recombinants, the smaller the map distance between two loci.
The usual prediction in F1 X F1 dihybrid crosses is 1:1:1:1 gametic ratios (eg 1/4 Mo+ Lav+, 1/4 Mo+ lav, 1/4 mo lav, 1/4 mo Lav+ as in the previous dihybrid example with lav (Lavender) and mo (mottled) heterozygotes), ie 25% for each gametes:
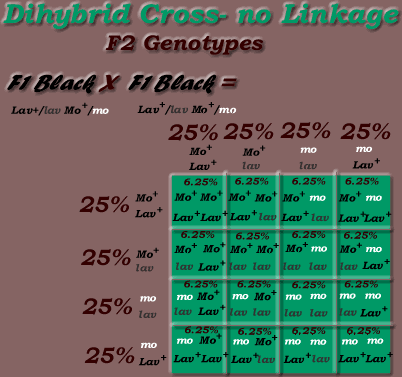
*Phenotypic Ratio = 9:3:3:1 (9/16 black, 3/16 black mottled, 3/16 lavender, 1/16 lavender mottled)
(56.25% black, 18.75% black mottled, 18.75% lavender, 6.25% lavender mottled)
-but the ratios with linked loci are different, depending on map distance, eg if the lavender locus & mottled locus had 10% linkage, & parental birds were P1: Mo+-Lav+ plus P2: mo-lav linkage, the F1 X F1 dihybrid crosses gametic ratios would be:
-45% Mo+ Lav+, 5% Mo+ lav, 45% mo lav, 5% mo Lav+
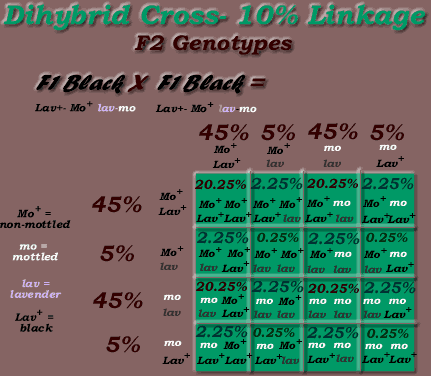
* Note, the above is an example only. The lav locus is NOT linked to the mo locus.
*Phenotypic % = 90 % P1 & P2 parental phenotypes, 10% crossovers
(70.25% black, 20.25% lavender mottled, 4.75% black mottled, 4.75% lavender)
Phenotypic ratio = 14:4:1:1
(14/20 black, 4/20 lavender mottled, 1/20 black mottled, 1/20 lavender)
A real example of close linkage is between the P (pea comb) locus from the O (Blue egg-shell), approximately 5 map units apart. Therefore, recombinants occur 5 % of the time. This is determined by crossing P-O P-O (pea-combed, blue egg-shell) X p+-o+ p+-o+ (single-combed, non-blue egg-shell) (or the opposite, crossing P-o+ P-o+ with p+-O p+-O), then breeding the F1 dihybrid offspring together. "Recombinants" is the total of both crossover segregates, ie both P/P o+/o+ (P-o+ P-o+) pea combed, non-blue egg-shell and p+/p+ O/O (p+-O p+-O) single combed, blue egg-shell) offspring (or with the second pairing, segregating F2 P-O P-O & p+-o+ p+-o+, as crossovers can occur both ways). So the number of single-combed blue egg-shell recombinants is half, ie 2.5% (2.5 in every 100 F2 offspring).
Sex-Linked, Linked LociIt is also possible for linkages to occur on the Z chromosome, ie both sex-linked & linked loci. As only males in avian species have a homologous set of sex chromosomes (ie, only males have two Z chromosomes), crossovers can only occur with males.
For example, if the mo & lav loci were on the Z chromosome (ie sex-linked), with a 10% linkage, the following results would occur:
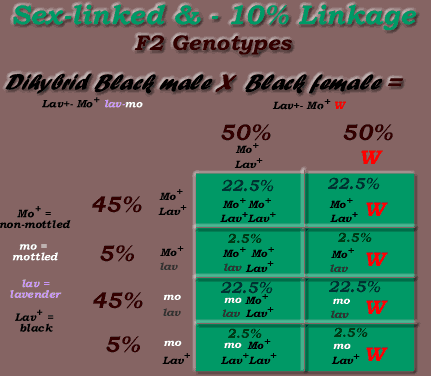
* Note, the above is an example only. The lav locus is NOT linked to the mo locus, & both are autosomes, NOT sex-linked.
Phenotypic Ratio = 95 % P1 & P2 parental phenotypes, 5% crossovers:
- 50% black males
- 22.5% black females, 22.5% lavender mottled females, 2.5% black mottled females, 2.5% lavender females
Once again, the crossover frequency for any two genes is determined from breeding results, but the fact that crossovers only occur with the males (with 2 Z chromosomes), this needs to be taken into consideration with the calculations. The daughters obtain their Z chromosome from the fathers, the reason why the crossovers are expressed in the females, with the above example. In the above example, since black mottled & solid lavender female offspring can only be produced if crossover occurs, you would record the number of this type offspring produced, divide this number by the total number of offspring, times by 100 (for percentage), then multiply this number by 2 to determine the total crossover rate in percentage. The female recombinant percentage is multplied by two, to obtain the total recombinant rate for both males & females:
eg, if 200 chicks hatched, with 5 females black mottled, 5 females solid lavender:
| Step 1: | 5 black mottled & 5 lavender females |
||
100 X |
-------------------------------- |
= 5 % recombinants |
|
200 offspring |
|||
//// |
/////// |
////////////////////////////////////////////// |
//////////////////////////////// |
| Step 2: | 5 X 2 |
= 10% total crossovers |
----------------------------------------------------------
Epistasis Dihybrid Inheritance Modes
Epistasis is where gene expression is affected by a gene from another locus. As noted above, usually the F2 offspring from a dihybrid pairing (genes not linked) segregate to the phenotypic ratio of 9:3:3:1. Where there is epistasis, this phenotypic ratio changes, eg:
- Dominant Epistasis: 12:3:1
- Recessive Epistasis: 9:3:4
* Therefore, the dihybrid ratios can be used to determine whether epistasis is affecting inheritance.
Dominant Epistatis Inheritance:
The following diagram is of Dominant Epistatis inheritance, ie E is epistatic - & masks expression of Co (hypostatic). The Columbian gene - Co (on its own - ie no Db, etc) is not expressed on the E, ER alleles (regardless that Co is a dominant gene), but Co expresses the Columbian phenotype on eWh, e+, eb, ey, etc alleles.
  |
* Note the above is not taking into consideration eumelanisers, nor incomplete dominance of E to eWh, etc, ie, the "black" phenotype in some may actually be closer to Brown Red/Grey phenotypes, etc depending on what other modifiers are present.
Recessive Epistatis Inheritance:
The following diagram is of Recessive Epistatis inheritance, ie Recessive white (c) is epistatic -masks all other plumage colours when homozygous, although a recessive gene:
  |
----------------------------------------------------------
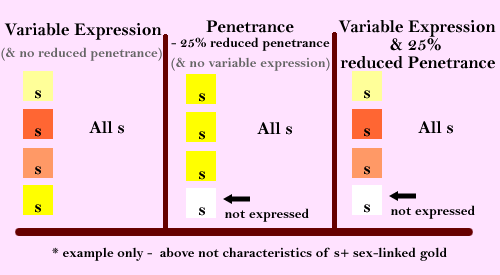
Variable Expressivity & Reduced Penetrance
These two are totally different concepts, with different statistical results. “Variable Expression” complies with expected inheritance ratios, & only relates to variance of phenotype in a bird, compared to his/her mates with the same genotype (eg light gold to dark gold variance, but all (100%) of birds with the same genotype expressing some shade of gold). “Penetrance” refers to how many (%) of the population that express the expected phenotype of a specific genotype, ie more to do with probabilities & the fact that some mutations don’t always express (eg some birds of the population with gold gene, not expressing any gold at all). Some mutations/genotypes have both variable expression & reduced penetrance.
Hopefully the following definitions & diagram will be clearer:
Definitions from “Genetics and Evolution of the Domestic Fowl” (by Lewis Stevens, 1991).
quote:
Expressivity:
The range of phenotypes expressed by a given genotype under any given set of environmental conditions.
Penetrance:
The proportion of individuals of a specified genotype that show the expected phenotype under a defined set of environmental conditions.
----------------------------------------------------------
\\// Home \\//Genetics Theory \\// Chicken Mutations\\// Chromosome Linkages\\// Chicken Variety Genotypes\\//
\\//Genetics Calculators\\// References/Links\\// About\\//